Abstract
The use of DNA-encoded libraries (DELs) has increased greatly over the last decade, and today a majority of pharmaceutical companies employ the technology. The technology may be applied to most soluble and purified targets. However, standard DEL technology has limitations; some targets are challenging to purify, and it is not possible to directly screen for cellular or biochemical activity. Numerous creative methods have been reported to overcome these limitations and expand DEL target scope. Reported proof-of-concept experiments include DEL selections of cell surfaces, and inside of living cells. Additional alternatives include the construction and biochemical screening of one-bead-one-compound (OBOC) DELs using picoliter aqueous droplets or micro-fabricated wells as containers. In these cases, the small-molecule moiety of the library member is liberated from its DNA barcode, and able to interact freely with the desired target. Lastly, patent literature suggests the ability to conduct cellular functional screens using OBOC DELs.
Graphical abstract

Keywords
DNA encoded libraries;High throughput screening;Hit discovery;Medicinal chemistry;Combinatorial chemistry
The popularity of DNA-encoded chemical library (DEL) technology has increased greatly over the last decade, and today a majority of pharmaceutical companies employ the technology to find hit chemical matter against soluble protein targets.1, 2 Generally, a DEL molecule consists of a DNA barcode covalently attached via a flexible linker to a small-molecule moiety. The barcode and small-molecule are constructed in a step-wise process employing split-and-pool combinatorial chemistry. In this manner, libraries of millions (or billions) of novel chemical structures are readily generated, with the caveat that the chemistry employed must be compatible with the presence of the encoding DNA. Additionally, byproducts and left-over starting materials cannot be purified away. After synthesis, the complex mixture of encoded chemical structures (the library) is then incubated with an immobilized protein target, and those library members which bind the target are captured, their DNA sequenced, and their corresponding chemical-structures thus ascertained (i.e. a DEL selection).
DEL selections may be applied to most any soluble and purified (usually protein) target. However, limitations exist when employing DELs as described above: i) it is not possible to conduct functional cellular or biochemical screens, ii) some targets are difficult to acquire in a soluble form, such as GPCRs and ion channels, iii) some targets lack a biologically relevant tertiary structure outside the cellular environment, for instance many transcription factors, iv) bacteria may be best screened directly, as cellular accumulation of the ligand is a key challenge to antibiotic discovery, v) novel chemical matter that breaks the rule-of-five (for instance macrocyclic peptides) is best screened in a cellular functional assay to assess both cell penetration and target engagement, and lastly vi) certain chemical classes (for instance molecules with covalent warheads) are arguably best assessed in a biochemical screen.
Numerous methods to overcome the limitations of DEL selections have been provided in the literature. Bear in mind that the true strength of DEL is the ability to rapidly synthesize new chemical structures. However, only femtomoles of each new chemical structure is produced, and so the goal of any new methodology is to make use of the miniscule quantities of encoded compounds that are synthesized.
DEL selections can be accomplished against targets overexpressed on the cell surface.3 The GPCR (NK3 tachykinin receptor) was overexpressed in HEK293 cells by transduction with a recombinant BacMam virus. Receptor expression levels on the cell surface were determined by a filter binding assay and 500′000 receptors per cell were detected; this high quantity of receptor was likely key for successful DEL screening. A negative control HEK293 cell line (lacking overexpression of NK3) was also prepared. A DNA-conjugated positive control (employing an analog of SB-235375, a known potent NK3 ligand) was synthesized. The DNA-conjugated positive control was observed to selectivity bind to the cell line overexpressing NK3 compared to the negative control cell line (determined by both fluorescence binding assays and DEL selection experiments followed by DNA sequencing).
Several DELs, totaling ~ 15 billion unique chemical structures, were used in the selection and a variety of novel chemotypes observed to enrich. Follow-up synthesis of compounds (lacking the DNA tag) were found to be highly active in a functional assay. For example, compound 1 (synthesized off-DNA) was found to inhibit NK3 with low nanomolar potency (Fig. 1). Compound 1 (on-DNA) was also shown by a cell sorting assay to bind cells overexpressing NK3. All the investigated compounds were highly selective for NK3 over NK1 or NK2. While the authors state that “The method is simple and broadly applicable to other GPCRs and integral membrane proteins”, we note that theirs remains the only successful report of such a selection using this technique.
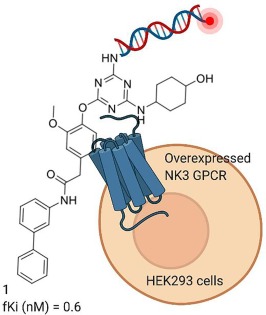
Fig. 1. DEL selection against a cell surface target. Traditionally, DEL molecules consist of a small-molecule ligand conjugated to a DNA barcode. Optionally, DNA-conjugated ligands possessing a fluorescent probe can be synthesized. The molecule shown was discovered from the cell surface DEL selection, and its activity measured by a functional FLIPR assay (fKi) and a 125I-NKB competition assay. (Created with BioRender.com).
A DEL selection methodology against membrane proteins on the surface of live cells that doesn’t require overexpression of the target protein was recently reported.4 Antibodies are used to selectively guide a DNA-conjugated phenylazide label to the target membrane protein, where crosslinking takes place upon ultraviolet irradiation (365 nm, 10 s). Three targets were separately selected against, folate receptor, carbonic anhydrase 12, and epidermal growth factor receptor. In each case, a 30 million member ssDNA DEL was incubated for 1 h with live HeLa cells following labeling of the desired membrane protein. Note that the label and the library members both possess a ssDNA oligomer, and these oligomers possess a complimentary region of 7 base pairs. Thus, the label reversibly hybridizes with library members and stabilizes the interaction of library members with the target protein (gain in stability is estimated to be ~6 kcal mol−1, corresponding to a ~ 20′000-fold boost in affinity). The cells were then subjected to 10 cycles of washing and resuspension. Library members bound to unlabeled proteins, and lacking the 6 kcal boost in affinity, were readily washed away. Bound library members were then eluted via heating the cells to 95 °C for 10 min. Hit molecules were discovered for all three investigated targets, suggesting that the technique is robust.
A proof-of-concept was published suggesting that a DEL selection could be conducted inside cells (Fig. 2).5 Halo-tag E. coli-dihydrofolate reductase (eDHFR) was expressed in FreeStyle 293-F cells. This particular cell line lacks the cGAS-STING pathway which detects cytosolic DNA, which may otherwise have interfered with the DEL selection. A DNA-conjugated positive control was synthesized using a trimethoprim analog as the ligand (reported off-DNA Kd = 1 nM). The DNA-conjugated positive control was further modified to display a sulfonyl fluoride moiety, a cell penetrating peptide (CPP) at the terminus of the DNA barcode, and optionally a 6-carboxyfluorescein amide (FAM) probe. The purpose of the CPP is to allow uptake of the DNA-encoded library members into the cell. A HaloTag-based chloroalkane penetration assay suggested some extent of endosomal escape and accumulation in the cytosol. Cellular uptake was also observed by fluoresce microscopy and shown to be dependent upon conjugation to the CPP.
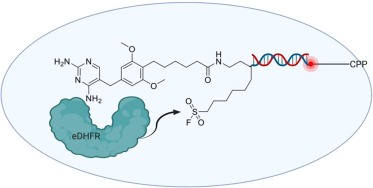
Fig. 2. Proof-of-concept for DEL selections inside live cells. The target Halo-tag E. coli-dihydrofolate reductase (eDHFR) was expressed in cells prior to incubation with a DNA-conjugated positive control. The DNA-conjugated positive controls were modified with a cell penetrating peptide (CPP) and a reactive moiety (in this case, a sulfonyl fluoride). Additionally, positive controls were synthesized possessing fluorescent probes and used to monitor cellular uptake. The small molecule moiety shown is an analog of trimethoprim, a previously reported ligand of eDHFR. The purpose of the reactive moiety is to covalently link the DNA barcode to the protein target. This concept was then used to select a 96 member DEL against Halo-tagged CBX7-ChD. (Created with BioRender.com).
The DNA-conjugated positive control (Fig. 2) was incubated with live cells prior to lysis and capture of Halo-tagged eDHFR. The experiment showed covalent modification of the captured eDHFR with the DNA-conjugated positive control. Covalent modification was dependent on the presence of the CPP, and the sulfonyl fluoride moiety; covalent modification did not occur in the presence of excess trimethoprim. Further experiments were conducted with Chromobox Protein Homologue 7 Chromodomain (CBX7-ChD), and the integral membrane protein δ opioid receptor (DOR). In all reported cases low to moderate enrichment of the DNA-conjugated positive control was observed relative to a DNA-conjugated (nonligand) negative control (following cell lysis and capture of the target protein via its Halo-tag). Note that recovery of DNA following cellular uptake was generally less than 10% as judged by qPCR, dependent on DNA length, and that a significant amount of that recovered DNA was likely trapped in endosomes. An extremely small focused library of 96 DNA-encoded conjugates was selected against Halo-tagged CBX7-ChD in live cells, cell lysates, and purified protein. Enrichment results were similar between live in-cell and lysate screens, and synthesized off-DNA molecules found to be potent in an orthogonal ligand displacement assay.
An alternative method of conducting a DEL selection inside of cells employs a single Xenopus leavis oocyte (Fig. 3).6 The oocyte is roughly 100′000 times larger than most other eukaryotic cells, with a volume of ~1 ul, and acts as a container for the DEL selection. Additionally, the oocyte is used to produce the protein target. Fused to the protein target is a ‘Prey’ protein. The purpose of the prey-bait system is to ensure that only DEL molecules bound to the desired target are later PCR amplified.
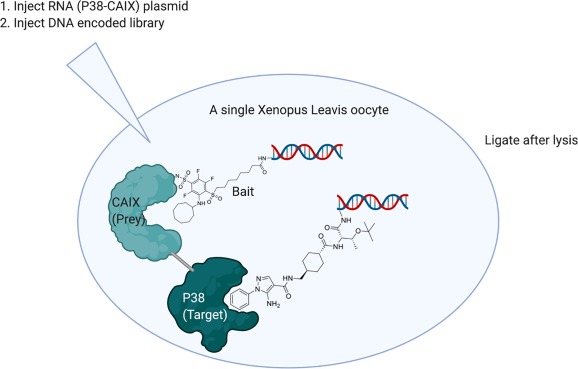
Fig. 3. A DEL selection inside a single oocyte. Before the selection, a fusion protein is expressed in the cell, consisting of target and a ‘Prey’ protein. DEL is then injected into the cell, along with a DNA-conjugated ligand known to bind the ‘Prey’ (referred to as ‘Bait’). After incubation, the cell is lysed, its contents diluted, and DNA ligase transiently added. If in close proximity, library members’ DNA will be ligated to Bait DNA, and the ligated DNA will be competent for later PCR amplification. (Created with BioRender.com).
To conduct a proof-of-concept selection, an oocyte was injected with mRNA coding for the expression of a CAIX-p38 fusion protein. Carbonic anhydrase 9 (CAIX) is the ‘Prey’ and p38 mitogen-activated protein kinase (p38) the target. Beforehand, a DNA-conjugated ligand which binds to the ‘Prey’ was synthesized (see ‘Bait’ in Fig. 3). Four days after injection of mRNA into the oocyte, 1012 molecules of DEL and 1011 molecules of ‘Bait’ were injected into the cell, and allowed to incubate for several hours. The single oocyte was then lysed, and the cytosolic contents diluted 10′000-fold in buffer prior to addition of T4 DNA ligase. Ligation was allowed to proceed for 4 mins prior to quenching. Only library members whose DNA is ligated to a ‘Bait’ molecule’s DNA is competent to then be amplified by PCR and sequenced.
The result of the in-cell p38 selection was similar to that obtained by a traditional cell-free selection against purified p38 protein; ~10 potential hit molecules were identified with either method and total enrichment levels were similar, and there was some overlap between enriched structures. By chance, one of the 10 molecules enriched in the in-cell selection was a known inhibitor of p38 with a reported IC50 of 500 nM (chemical structure provided in Fig. 3).
The ability to interrogate DELs employing a functional read-out likely requires that each library member be assayed individually for biochemical or cellular activity. Further, the ability to assess a library member’s chemical moiety’s ability to penetrate a living cell requires, at some point, that there be no covalent connection between DNA barcode and ligand. One way to accomplish this is to synthesize one-bead-one-compound (OBOC) DELs using split-and-pool combinatorial chemistry (in the same manner as solution phase libraries).7 The small-molecule ligand can be synthesized step-wise from the terminus of a UV (Fig. 4A) or enzyme (Fig. 4B) labile linker. The encoded small-molecule can then be released from the bead following coencapsulation with target inside an aqueous droplet.
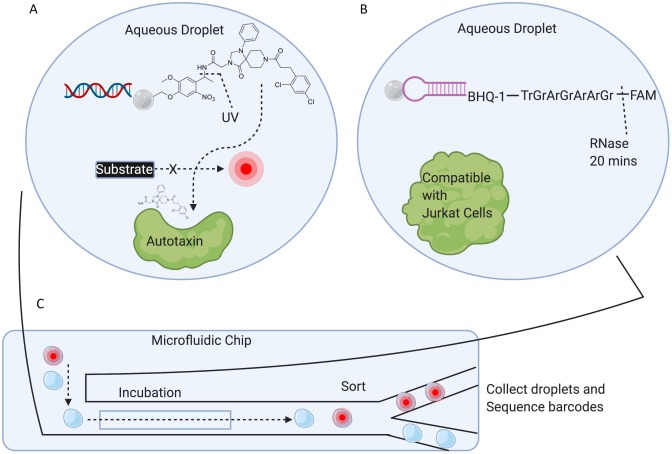
Fig. 4. One-bead-one-compound (OBOC) DELs with labile linkers. A) Biochemical DEL screen against Autotaxin using an OBOC DEL. The chemical structure of the positive control bead, used to optimize the screening conditions, is shown. Library members consisted of a bead labeled with DNA (the barcode) and a small-molecule moiety connected to the bead via a photolabile linker. Library beads, Autotaxin, and self-quenching substrate were coencapsulated in droplets, in a manner such that few droplets had more than 1 bead. B) A potential method of constructing an OBOC DEL consists of attaching the small-molecule moiety to the terminus of an RNA strand. The small-molecule can then be released from the bead following incubation with RNase. Coencapsulation is reported to be compatible with cell viability. Note that only a single test-case library member was synthesized and investigated. C) Beads and target are coencapsulated within custom made microfluidic chips, followed by incubation and finally sorting of droplets according to fluorescence (or fluorescence anisotropy). (Created with BioRender.com).
In a proof-of-concept, an OBOC DEL of ~70′000 members was screened against Autotaxin employing a self-quenching artificial substrate that would fluoresce after cleavage by the active enzyme (Fig. 4A).7 Hence, droplets that remained non-fluorescent were assumed to contain potent Autotaxin inhibitors. Positive control beads were made employing a known Autotaxin inhibitor (chemical structure shown in Fig. 4A). These positive control beads were used to optimize the assay conditions, and prove that photorelease of the ligand is required for enzyme inhibition. An integrated microfluidic chip was used to screen the DEL: droplets were formed, dosed with UV, incubated, and finally sorted based on droplet fluorescence all within the custom-made chip. The flow rate of droplets through the chip allowed for the sampling of ~200′000 library beads. The DNA attached to the beads (from the collected non-fluorescent droplets) was then amplified and sequenced, and 35 molecules were deemed as enriched. Five of those molecules were chosen for resynthesis, and all 5 were found active in a follow-up biochemical assay.
A second proof-of-concept OBOC DEL screen was accomplished targeting the receptor tyrosine kinase discoidin domain receptor 1 (DDR1).8 The DDR1 screen used the same ~70′000 member OBOC DEL as used in the above Autotaxin screen. The screen was a competition assay displacing a fluorescently labeled DDR1 probe. Droplet sorting was based on fluorescence anisotropy, and the entire process (droplet formation to sorting) took place inside a custom made microfluidic chip. Approximately 1 million beads were screened. After bead collection, DNA amplification, and sequencing, roughly 300 library molecules were deemed to enrich, many of whose small-molecule moieties were structurally identical or similar to previously reported DDR1 inhibitors.
As illustrated in Fig. 4B, the small-molecule can also be connected to the bead through a region of ssRNA, and released from the bead by the addition of RNase.9 This cleavable linker system was investigated by generating a control bead possessing a quencher and fluorophore separated by a 7 bp ssRNA linker, and RNase cleavage determined complete in ~20 min following coencapsulation. Additionally, the system was shown to be compatible with coencapsulation of Jurkat cells. Note that no actual screen using this ssRNA linkage was reported, however the design does provide a potential alternative to the previously reported photolabile linker.
Potential methods of screening OBOC DELs that avoid using custom-made microfluidic chips and droplet sorters have been suggested. (Note that while some experimental details are reported, complete proof-of-concept experiments are not provided). Such methods would avoid the burden of producing custom chips. Also, the numeric size of OBOC libraries that can be screened is limited by the frequency of droplet sorting. In one example (Fig. 5A), bead and bacteria are coencapsulated in a droplet, and fluoroquinolone antibiotic released from the bead upon photodosing.10 Ensuing bacterial death results in the leakage of endonucleases and destruction of the bead’s DNA barcodes. Beads are collected following incubation with bacteria, all remaining DNA amplified and sequenced, and those barcodes deemed missing then ascribed to cytotoxic small-molecules. In a similar test case, a BstC1 endonuclease site is placed on the DNA barcode of the bead, between a iFluorT (fluorescein dT modification) and a quencher.11 In this case, droplets become fluorescent after coencapsulation of bead and BstC1, and alternatively remain non-fluorescent when coencapsulated with an inhibitor of BstC1. In addition, the alteration of the DNA barcode attached to the bead potentially allows for selective PCR amplification, and thus droplet sorting is potentially not required.
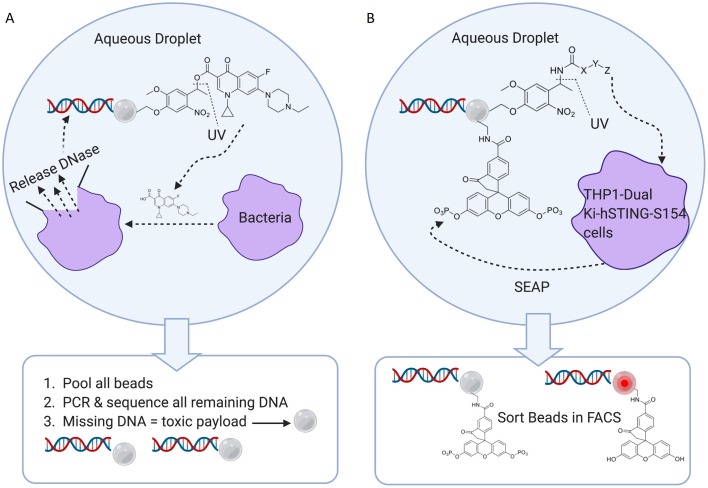
Fig. 5. Alternative methods for potential OBOC DEL screens. A) Antibiotics lead to bacterial death and release of endonucleases that destroy the DNA barcode. Thus, droplet sorting is unnecessary. B) Cellular release of SEAP is used to measure compound activity, as phosphatase de-quenches a bead-bound fluorescein derivative. Following incubation of bead and cells in droplets, the beads can be collected and sorted on a fast and robust commercial cell sorter. (Created with BioRender.com).
In another reported scheme (Fig. 5B), beads and live cells are encapsulated in droplets, and library members released upon photodosing.10 In this instance, the cell line expresses secreted alkaline phosphatase (SEAP) constitutively. The secreted phosphatase unquenches a fluorescein derivative (1-oxo-3′,6′-diphosphonooxy-spiro[isobenzofuran-3,9′-xanthene]-5-carboxylic acid) attached to the bead. If the photocleaved library member enters the cell and inhibits the STING protein, then SEAP is not expressed. Following incubation, all beads can be collected and sorted on a commercial cell sorter. The method has the potential of avoiding the use of custom chips, and replaces the use of droplet sorters with faster and commercially available cell sorters. (Again, note that while some experimental details are reported, complete proof-of-concept experiments are not provided).
The examples of OBOC DEL screening described above require coencaspulation of beads with target in an aqueous droplet. An alternative is the use of custom picowells as illustrated in Fig. 6 (a plastic microwell array may possess 800′000 wells).12 The picowells can be coated with Pluronic 127 to provide a hydrophilic surface, followed by a coating of vitronectin on the bottom of the wells such that cells can adhere to the bottom of the picowells. OBOC DEL may be added to the top of the plate in excess to the number of picowells, and after a brief centrifugation, the beads can form a ‘cap’ atop each picowell. The wells are ~ 12 picoliters in volume, and swollen beads are 10–14 µm in diameter. In a reported example, HeLa cells are transfected with IKZF1-GFP, and positive control beads are loaded with a lenalidomide analog connected via a UV labile linker. Release of the lenalidomide analog results in degradation of the IKZF1-GFP fusion protein (presumably via a cereblon mediated ubiquitination process) and loss of fluorescence signal. It’s unclear exactly how the results of such a cellular functional screen would be measured. A partial proof-of-concept experiment is detailed however, where it’s shown that DNA barcodes can be directly sequenced from the beads as they sit atop the picowells. There are reported to be 60 million copies of the DNA barcode per bead, and the hairpin is designed in such a manner that the 3′-terminal nucleotides of the hairpin serves as a sequencing primer (not shown in Fig. 6). Note that the picowells themselves can also be directly assayed for fluorescence. Lastly, the beads can be collected from the picowells if desired; polyacrylamide can be poured over the plates, allowed to harden, and then removed with all the beads attached in the proper order.
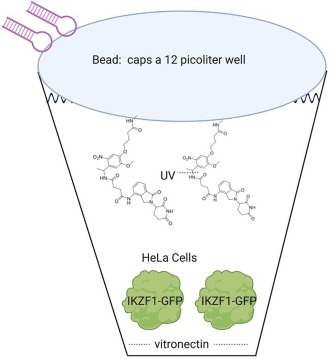
Fig. 6. Picowells may be used to co-incubate beads with target proteins or cells. (Created with BioRender.com).
The advantages of each of the above discussed DEL interrogation methods is listed in Table 1. Cell surface DEL selections potentially allow for the discovery of ligands that bind the extracellular domains of membrane proteins. The recent publication by Huang et al.4 provides encouragement that this paradigm may be robust enough for use in the pharmaceutical industry. Note that membrane proteins have also been solubilized, purified, and investigated using standard DEL selection protocols.13, 14 However, the isolation of membrane bound targets in the functional state, purity, and concentration required for a DEL selection is challenging. The use of thermo-stabilized mutants for DEL selections appears promising (as was done for the GPCR protease-activated receptor 2).13 However, unfortunately, thermo-stabilized membrane proteins are not commonly available.
Table 1. Summary of reported functional and cellular DEL screening methods.
| Interrogation Method | Figure | Advantages | Limitations | Proof-of-concept detailed |
|---|---|---|---|---|
| Cell surface affinity selection | 1 |
|
|
Yes |
| In-cell affinity selection | 2–3 |
|
|
Yes |
| OBOC DEL biochemical Screen | 4 |
|
|
Yes |
| OBOC DEL cellular functional screen | 5–6 |
|
|
No |
In-cell affinity selections (Fig. 2, Fig. 3) slightly improves DEL target scope by allowing for selections against difficult to purify proteins under physiologically relevant conditions.5, 6 However, the technique described by Hansen et al.6 is limited to cytosolic soluble proteins, and is not applicable to DNA binding proteins (such as transcription factors). Additionally, the technique lacks the ability to screen for the cellular penetration of ligands, as the library is injected directly into the cell.
OBOC DEL biochemical screens provide an alternative to affinity based DEL selections (Fig. 4).7, 8 OBOC small-molecule moieties are released from their beads upon screening, and thus are free to interact with their target without interference from a covalently attached DNA barcode. For some targets, only biochemically active hit molecules are desired, and so directly assaying for such activity may be preferable. There are further differences between a DEL affinity selection and a DEL biochemical screen. For a DEL affinity selection, high quality protein is used in excess, and minute compound impurities in the library are unlikely to bring about false positives. For a DEL biochemical screen, the ligand must be in excess of the protein, and protein purity should be less important. Also, as true for all biochemical assays, minute quantities of highly potent impurities could result in false positives. Interrogating a large number of different DEL molecules via a DEL biochemical screen is more difficult as compared to a DEL affinity selection. Each library member in a DEL screen must be contained in a single aqueous droplet and successively sorted7, or placed in separate nanowells12 and individually assessed. Note that Cochrane et al.7 screened a relatively small DEL of ~70′000 members. However, potential methods to increase throughput have been suggested including the use of faster commercial cell sorters (Fig. 5b) or methods that eliminate the need for any sorting (Fig. 5a). Looking forward, we presume that the pharmaceutical industry will screen focused DELs (numeric size of less than a million) designed around chemical fragments of interest, in an effort to increase potency and/or selectivity.
OBOC DEL cellular functional screening (Fig. 5, Fig. 6) would allow for the rapid screening of novel chemical matter for both cellular penetration and target engagement. Additionally, such screens would potentially increase DEL target scope to include pathways, large biomolecular complexes, transmembrane proteins, and difficult to purify proteins. Cellular screens would also be advantageous when interrogating chemical space that is known to have difficulties penetrating cells (such as macrocyclic peptides or small-molecules conjugated to cargo such as antisense oligonucleotides), or for particular therapeutic areas where cellular accumulation of ligand is of key importance such as antibacterial screening. However, while schemes for DEL cellular functional screens have been reported, no reproducible proof-of-concept has to-date been detailed.9–12 Note that DEL cellular screens share many of the same strengths and weaknesses as discussed above for DEL biochemical screening.
As discussed herein, numerous research groups have tackled the limitations of current DEL selections in various and creative ways. The field has developed quite recently: most of the work discussed in this digest was published in the last two years, and only a single review on this topic has been previously published.15 Importantly, there has been some recent success, and publication of detailed proof-of-concept experiments (Table 1). Further research is clearly underway in numerous academic and industrial labs, and an expectation of continued scientific breakthroughs over the next few years appears reasonable.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.